@spiritnotes
2016-03-07T03:53:16.000000Z
字数 3612
阅读 16408
机器学习实践 -- breast cancer数据集
机器学习实践
获取
sklearn数据集中已经包含该数据,可以直接获取。
cancers = datasets.load_breast_cancer()
清理
数据一共有569组30维。其中两个分类分别为
| 类型 | 个数 |
|---|---|
| 良性 benign | 357 |
| 恶性 malignant | 212 |
从数据中看无空值,有几个属性的最小值为0。
cancers_pd.isnull()cancers_pd.min(axis=0)
分别是
mean concavity 0mean concave points 0worst concavity 0worst concave points 0
这几个指的是凹度和凹点,是可以为0的。从数据的直方图上也可以看出,其分布是相对连续的。
分析
PCA降维
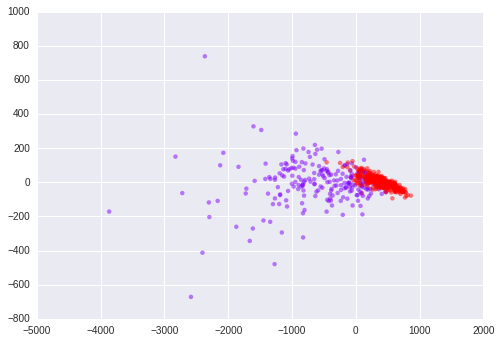
from sklearn.decomposition import PCApca = PCA(n_components=2)data_pca = pca.fit_transform(data)plt.scatter(data_pca[:, 0], data_pca[:, 1], c=target, edgecolor='none', alpha=0.5, cmap=plt.cm.get_cmap('rainbow', 2))
将数据按照PCA降维后,可以看到其两种数据还算比较容易区分。
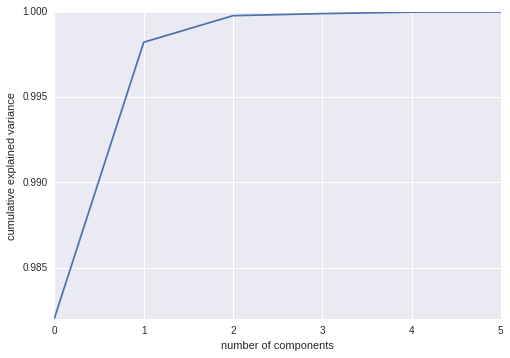
从能量图中也可以看到,前几维就可完全代表数据。
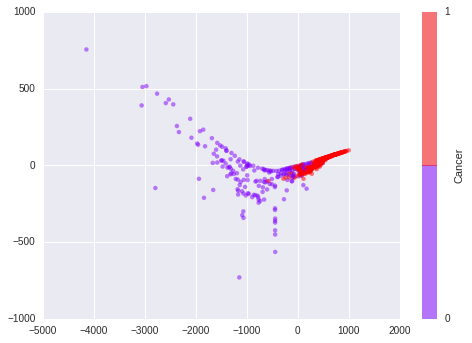
IsoMap
from sklearn.manifold import Isomapiso = Isomap(n_components=2)data_projected = iso.fit_transform(data)plt.scatter(data_projected[:, 0], data_projected[:, 1], c=target,edgecolor='none', alpha=0.5, cmap=plt.cm.get_cmap('rainbow', 2));
violinplot
for column_index, column in enumerate(data_sk.feature_names):if column_index%4 == 0:plt.figure()plt.subplot(2, 2, column_index%4 + 1)sb.violinplot(x='cancer', y=column, data=data_pd)
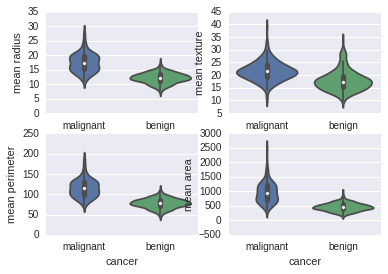
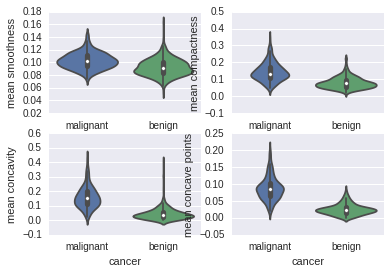
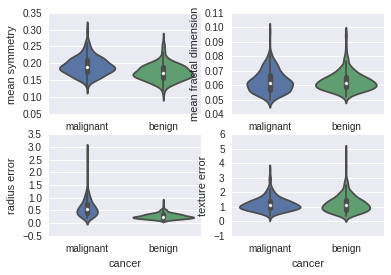
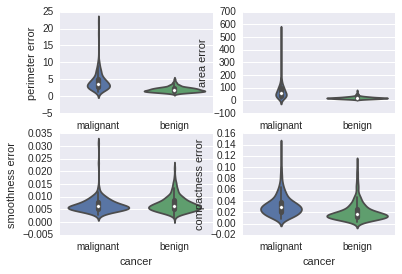
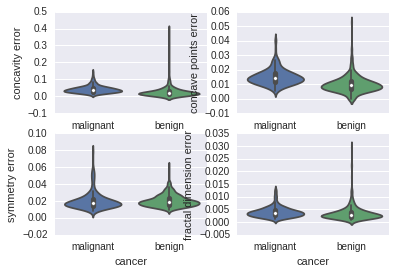
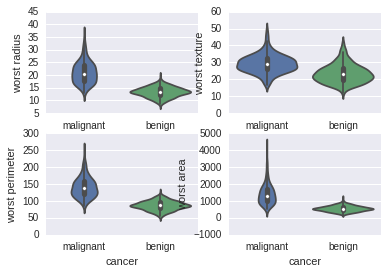
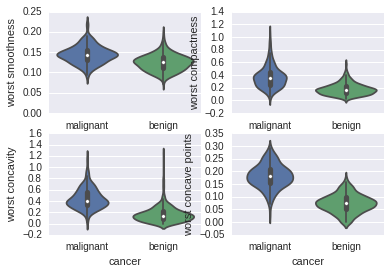
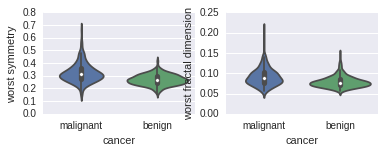
从上面图可以看到如下的序列号两者相差较大
[0, 2, 3, 6, 7, 10, 12, 13, 25, 26, 27]
pairplot

从pairplot图中可以看到其中有部分属性是线性相关的。另外从数据中可以看到两种Cancer之间没有明显的区分,从很多图上可以看到是有少许数据重合。
分类器训练
KNN
from sklearn.neighbors import KNeighborsClassifierclf = KNeighborsClassifier()
通过GridSearchCV可得其最佳参数如下
(0.93145869947275928,KNeighborsClassifier(algorithm='auto', leaf_size=30, metric='minkowski',metric_params=None, n_jobs=1, n_neighbors=10, p=2,weights='uniform'),{'n_neighbors': 10, 'weights': 'uniform'})
逻辑回归
from sklearn.grid_search import GridSearchCVfrom sklearn.linear_model import LogisticRegressionclf = LogisticRegression(penalty='l2')C = [0.1, 0.5, 1, 5, 10]param_grid = [{'C': C}]grid_search = GridSearchCV(clf, param_grid=param_grid, cv=10)grid_search.fit(data, target)
运行结果为
(0.95079086115992972,LogisticRegression(C=0.5, class_weight=None, dual=False, fit_intercept=True,intercept_scaling=1, max_iter=100, multi_class='ovr', n_jobs=1,penalty='l2', random_state=None, solver='liblinear', tol=0.0001,verbose=0, warm_start=False),{'C': 0.5})
SVM
其结果为
(0.95430579964850615, SVC(C=1, cache_size=200, class_weight=None, coef0=0.0,decision_function_shape=None, degree=3, gamma='auto', kernel='linear',max_iter=-1, probability=False, random_state=None, shrinking=True,tol=0.001, verbose=False), {'C': 1, 'kernel': 'linear'})
决策树
结果为
(0.9472759226713533,DecisionTreeClassifier(class_weight=None, criterion='entropy', max_depth=15,max_features=None, max_leaf_nodes=None, min_samples_leaf=8,min_samples_split=3, min_weight_fraction_leaf=0.0,presort=False, random_state=None, splitter='best'),{'criterion': 'entropy','max_depth': 15,'min_samples_leaf': 8,'min_samples_split': 3})
随机森林
结果为
0.96836555360281196,RandomForestClassifier(bootstrap=True, class_weight=None, criterion='gini',max_depth=None, max_features='auto', max_leaf_nodes=None,min_samples_leaf=1, min_samples_split=2,min_weight_fraction_leaf=0.0, n_estimators=200, n_jobs=1,oob_score=False, random_state=0, verbose=0, warm_start=False),{'n_estimators': 200})
汇总
| Classifier | score | std |
|---|---|---|
| KNN | 0.931 | 0.033 |
| LogisticRegression | 0.950 | 0.018 |
| SVC | 0.954 | 0.019 |
| DecisionTreeClassifier | 0.947 | 0.026 |
| RandomForestClassifier | 0.968 | 0.025 |
提升真阳率
采用最佳的逻辑回归参数以及随机的0.2测试集划分,可得正确率 92.9%。混淆矩阵如下:
from sklearn.cross_validation import train_test_splitXtrain, Xtest, ytrain, ytest = train_test_split(data, target, test_size=0.2,random_state=2)clf.fit(Xtrain, ytrain)clf.score(Xtest, ytest)
| 实际\预测 | P | N |
|---|---|---|
| P | 64 | 5 |
| N | 3 | 42 |
计算出来,假阳率为3/45=0.067,真阳率为64/69=0.928。逻辑回归的predict_proba输出为分别为良性和恶性的概率。我们取predict_proba1/predict_proba[0]作为实际的手动判断标准。则其ROC图如下:
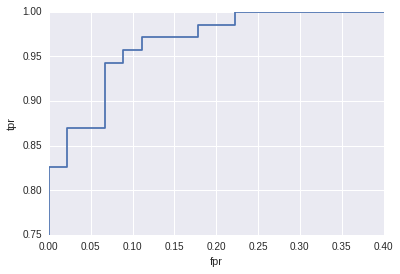
如果希望将真阳率提高到97%,则根据查表可得阀值为0.45。因此修改判决方法为
new_pre = (clf.predict_proba(Xtest)[:,1]/clf.predict_proba(Xtest)[:,0]>0.45)*np.ones(len(Xtest))
| 实际\预测 | P | N |
|---|---|---|
| P | 67 | 2 |
| N | 5 | 40 |
计算出来,假阳率为5/45=0.111,真阳率为67/69=0.971
代码
GitHub: https://github.com/spiritwiki/codes/tree/master/data_breastcancer
