@agpwhy
2021-10-16T05:22:01.000000Z
字数 2919
阅读 2040
王胖的生信笔记第21期:网友的irGSEA包
前段时间看到一个优秀的网友自己学生信到做出了一个优秀的单细胞测序数据分析整合工具包irGSEA(https://chuiqin.github.io/irGSEA/index.html),现在借着这个机会把这个教程跟着跑一下给大家分享一下。
这个包有些依赖的部分可能需要R升级到最新的版本(4.1.1),否则的话,bubbleplot那个可能会跑出问题。因为这个原因,我总算把拖了很久的老版本的R升级了,非常花时间。但是在热心网友的指点下,获得了一些比较节约时间的方法,下一期给大家分享。关于这个包,作者也在生信技能树上有投稿,欢迎大家去阅读。
GSEA
GSEA其实是一种富集方式,是在设置cutoff之外,给予排名后,再加以富集;比起单纯的富集,可能更能把一些微妙的差异体现出来。当然具体的原理还是要去学习数学,这里我就不献丑了。不过这里用的不仅是GSEA(其实是ssGSEA,即单样本GSEA),还有singscore,AUCell,UCell共四种方式。最后得到的结果是通过多种基因集富集方法分别对单个细胞进行打分,并生成多个基因集富集分数矩阵,然后通过RRA(秩聚合算法,我也不太理解这个数学原理)进行综合评估,而非简单的取交集得到在多种富集方法中综合考虑下来显著富集的基因集。
安装准备
cran.packages <- c("msigdbr", "dplyr", "purrr", "stringr","magrittr","RobustRankAggreg", "tibble", "reshape2", "ggsci", "tidyr", "aplot", "ggfun", "ggplotify", "ggridges", "gghalves", "Seurat", "SeuratObject", "methods", "devtools", "BiocManager","data.table","doParallel", "doRNG")
if (!requireNamespace(cran.packages, quietly = TRUE)) {
install.packages(cran.packages, ask = F, update = F)
}
bioconductor.packages <- c("GSEABase", "AUCell", "SummarizedExperiment", "singscore", "GSVA", "ComplexHeatmap", "ggtree","Nebulosa")
if (!requireNamespace(bioconductor.packages, quietly = TRUE)) {
BiocManager::install(bioconductor.packages, ask = F, update = F)
}
if (!requireNamespace("UCell", quietly = TRUE)) {
devtools::install_github("carmonalab/UCell")
}
if (!requireNamespace("irGSEA", quietly = TRUE)) {
devtools::install_github("chuiqin/irGSEA")
}
如果最后devtools::install_github有问题,欢迎学习使用yulabs.utils工具包。如果查下来还不会,欢迎私信交流。
搞一个公共数据尝试
示范教材里用的是pmbc3k.final的公共数据,我们这里换一个新的。GSE159929(这个公共数据集还是个好东西,欢迎大家探索)里面肝的数据。如果不会下载或者有需要欢迎私信交流。
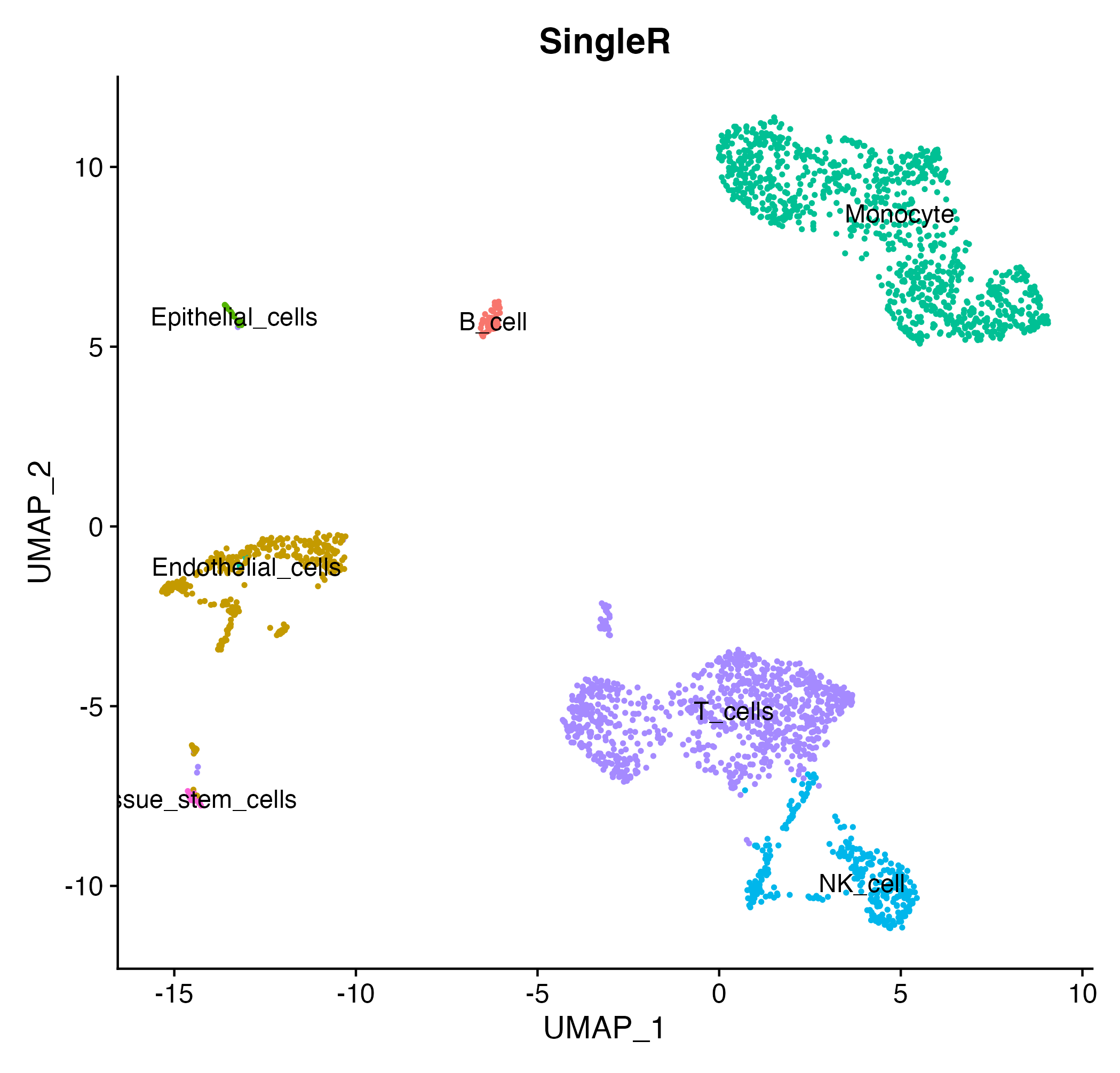
scRNA <- UpdateSeuratObject(scRNA)
DimPlot(scRNA, reduction = "umap",group.by = "SingleRlabel",label = T) + NoLegend()
library(UCell)
library(irGSEA)
scRNA <- irGSEA.score(object = scRNA, assay = "RNA",slot = "data", seeds = 123, ncores = 1,min.cells = 3, min.feature = 0,custom = F, geneset = NULL, msigdb = T,species = "Homo sapiens", category = "H",subcategory = NULL, geneid = "symbol",method = c("AUCell", "UCell", "singscore","ssgsea"),aucell.MaxRank = NULL, ucell.MaxRank = NULL,kcdf = 'Gaussian')
到这一步,四种方式的富集就计算好了,但是还需要整合
result.dge <- irGSEA.integrate(object = scRNA,group.by = "SingleRlabel",metadata = NULL, col.name = NULL,method = c("AUCell","UCell","singscore","ssgsea"))
到这里就算准备好了。
开始可视化
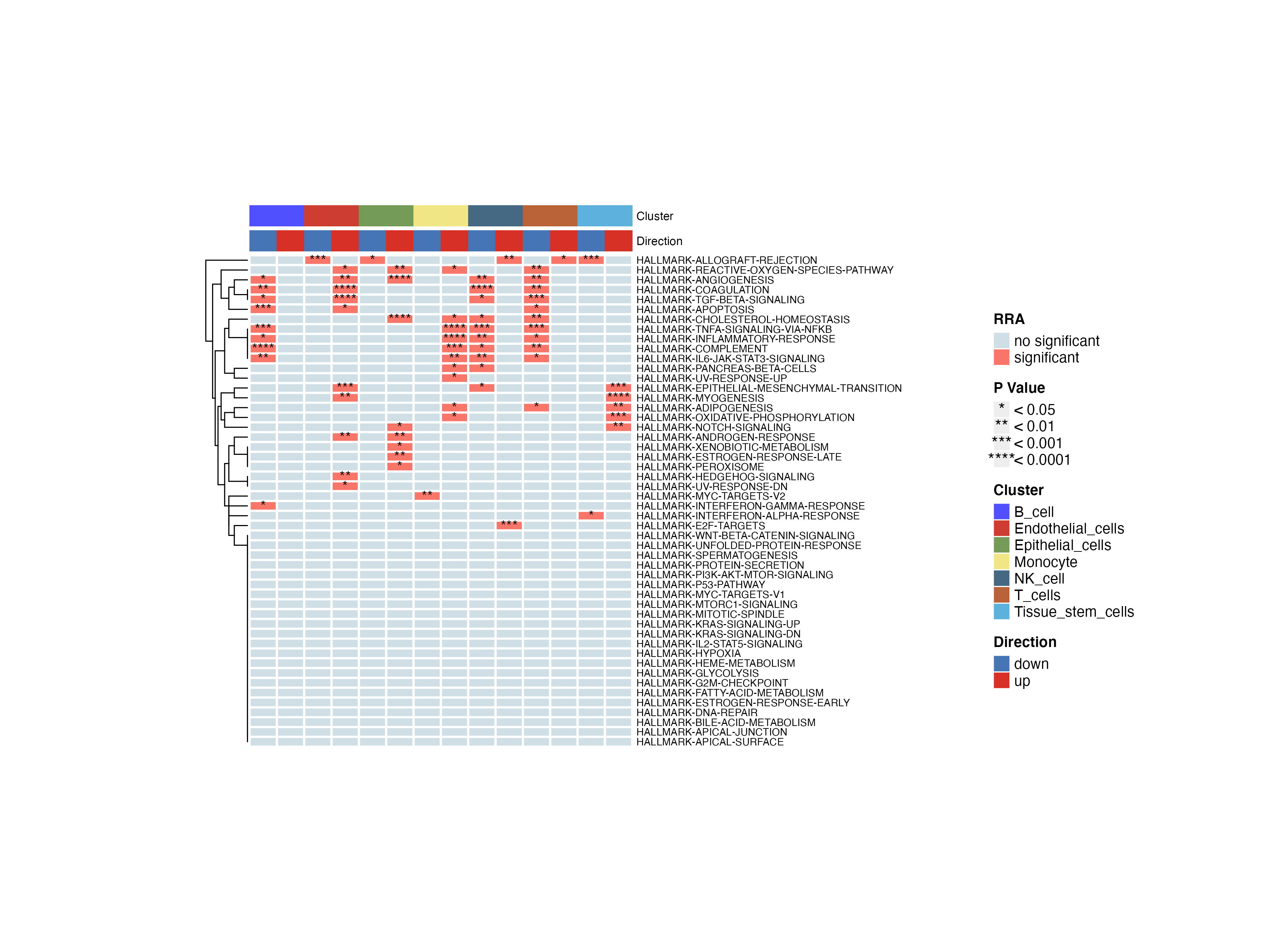
irGSEA.heatmap.plot <- irGSEA.heatmap(object = result.dge,
method = "RRA",
top = 50,
show.geneset = NULL)
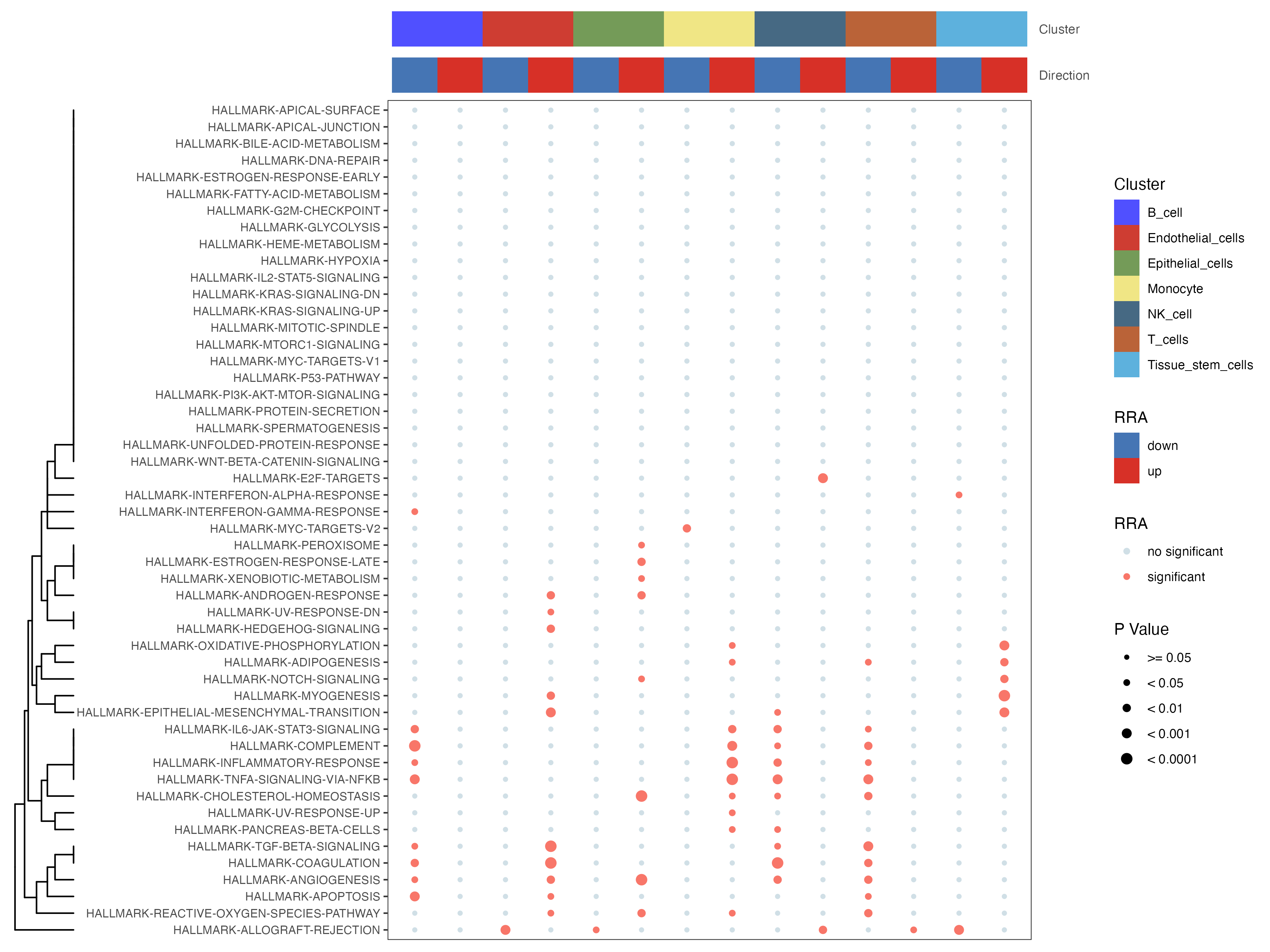
irGSEA.bubble.plot <- irGSEA.bubble(object = result.dge,method = "RRA",top = 50)
irGSEA.upset.plot <- irGSEA.upset(object = result.dge,method = "RRA")
irGSEA.barplot.plot <- irGSEA.barplot(object = result.dge,
method = c("AUCell", "UCell", "singscore",
"ssgsea"))
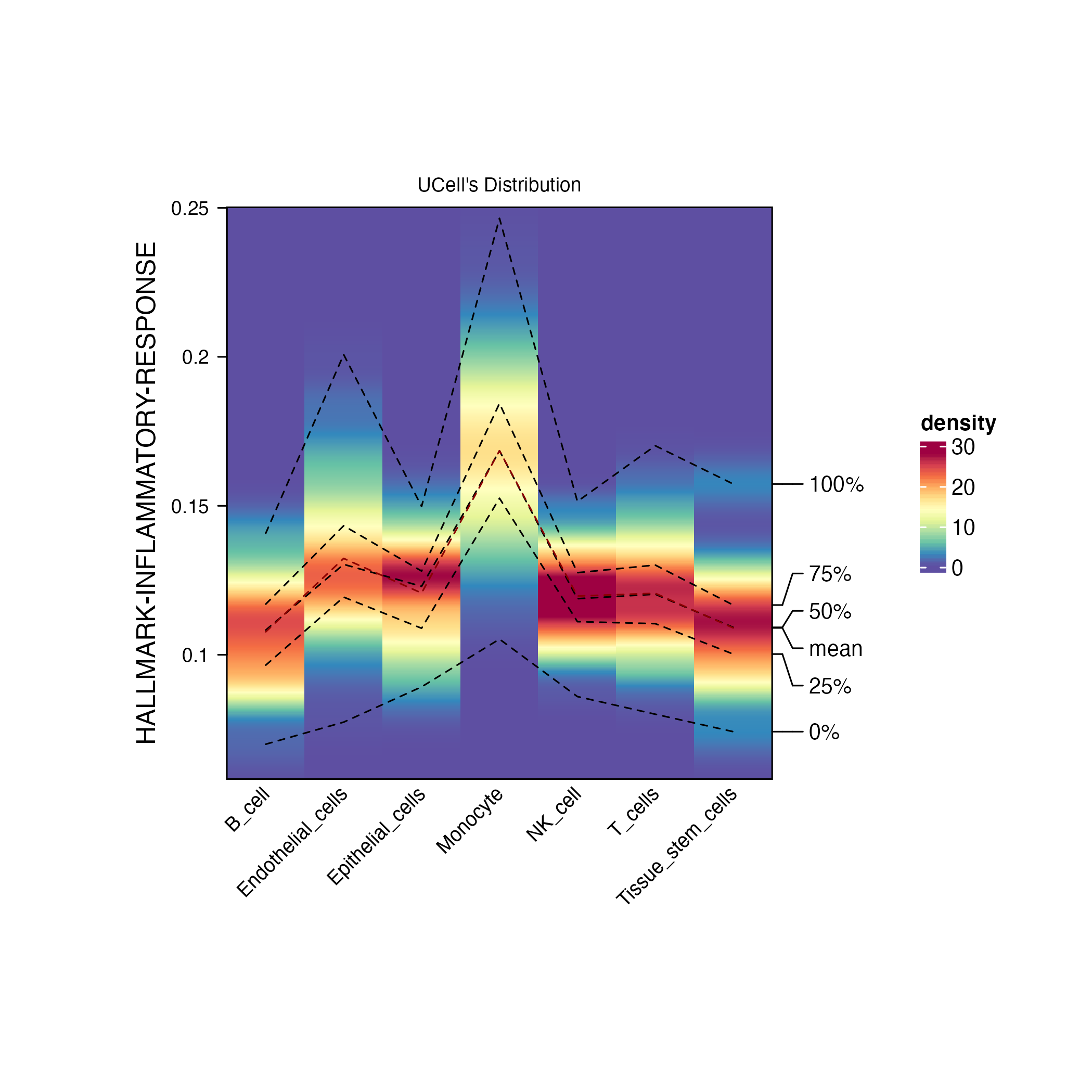
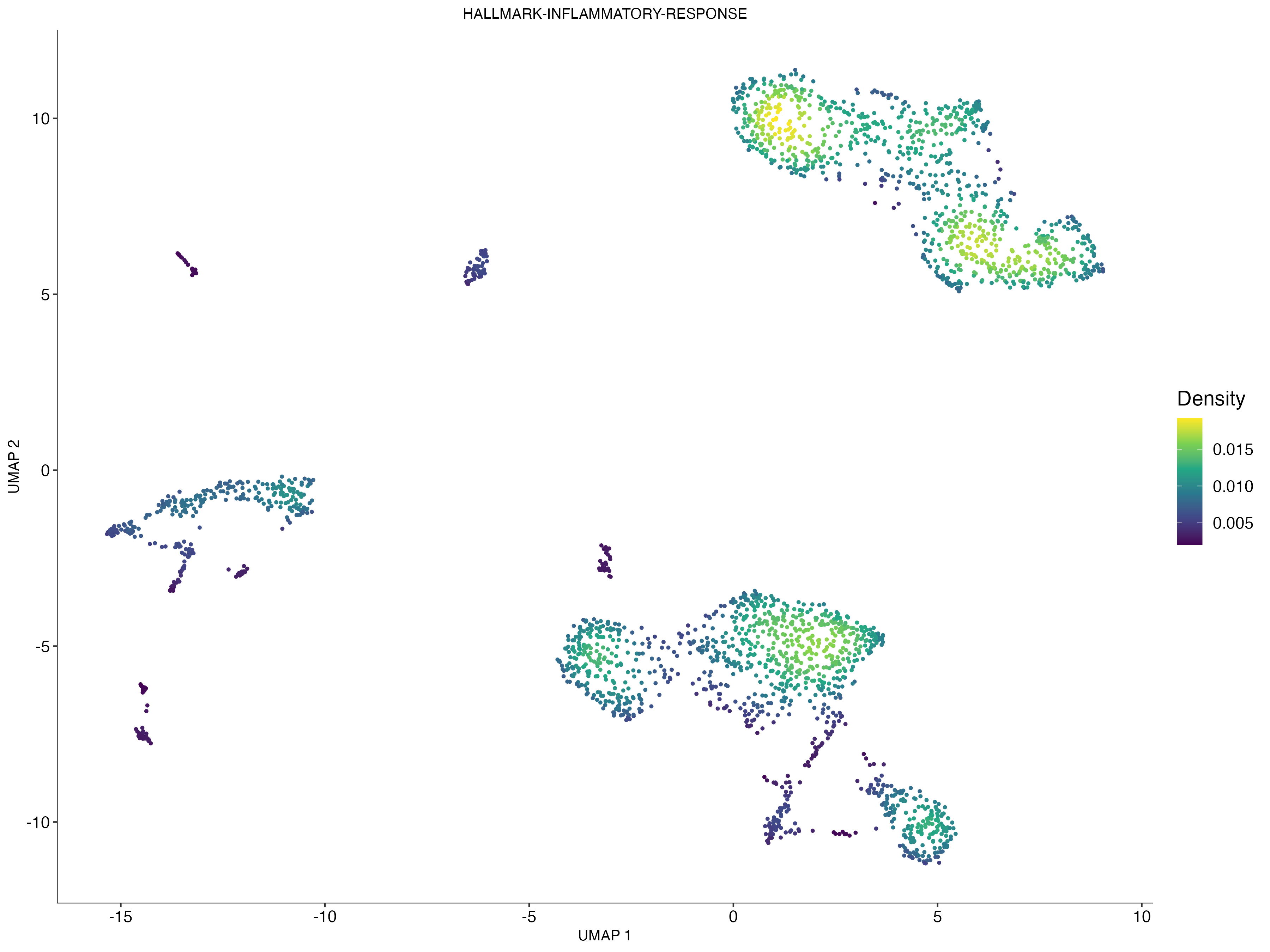
scatterplot <- irGSEA.density.scatterplot(object = scRNA,method = "UCell",show.geneset = "HALLMARK-INFLAMMATORY-RESPONSE",reduction = "umap")
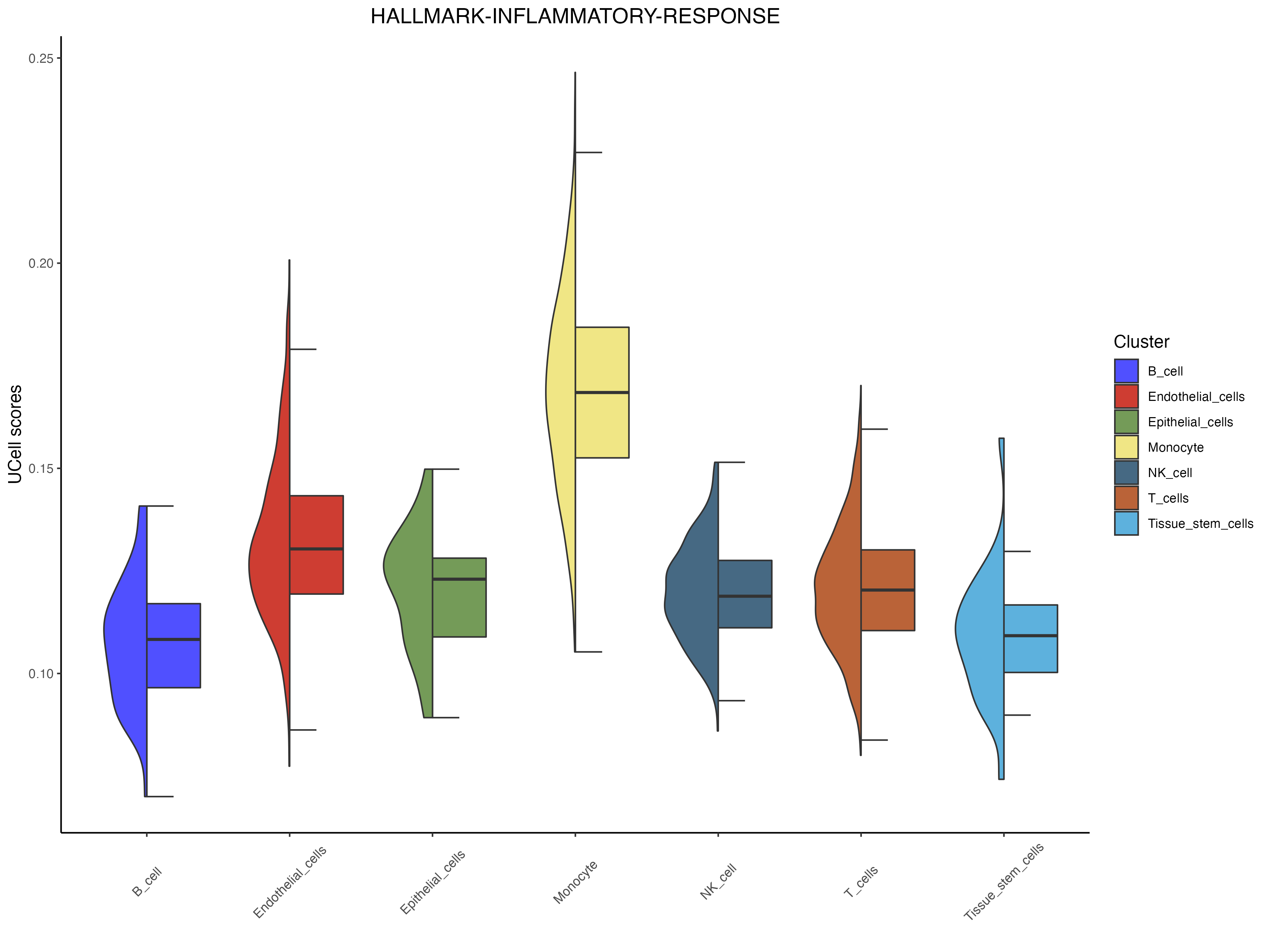
`halfvlnplot <- irGSEA.halfvlnplot(object = scRNA,method = "UCell",show.geneset = "HALLMARK-INFLAMMATORY-RESPONSE")
ridgeplot <- irGSEA.ridgeplot(object = scRNA,method = "UCell",show.geneset = "HALLMARK-INFLAMMATORY-RESPONSE")
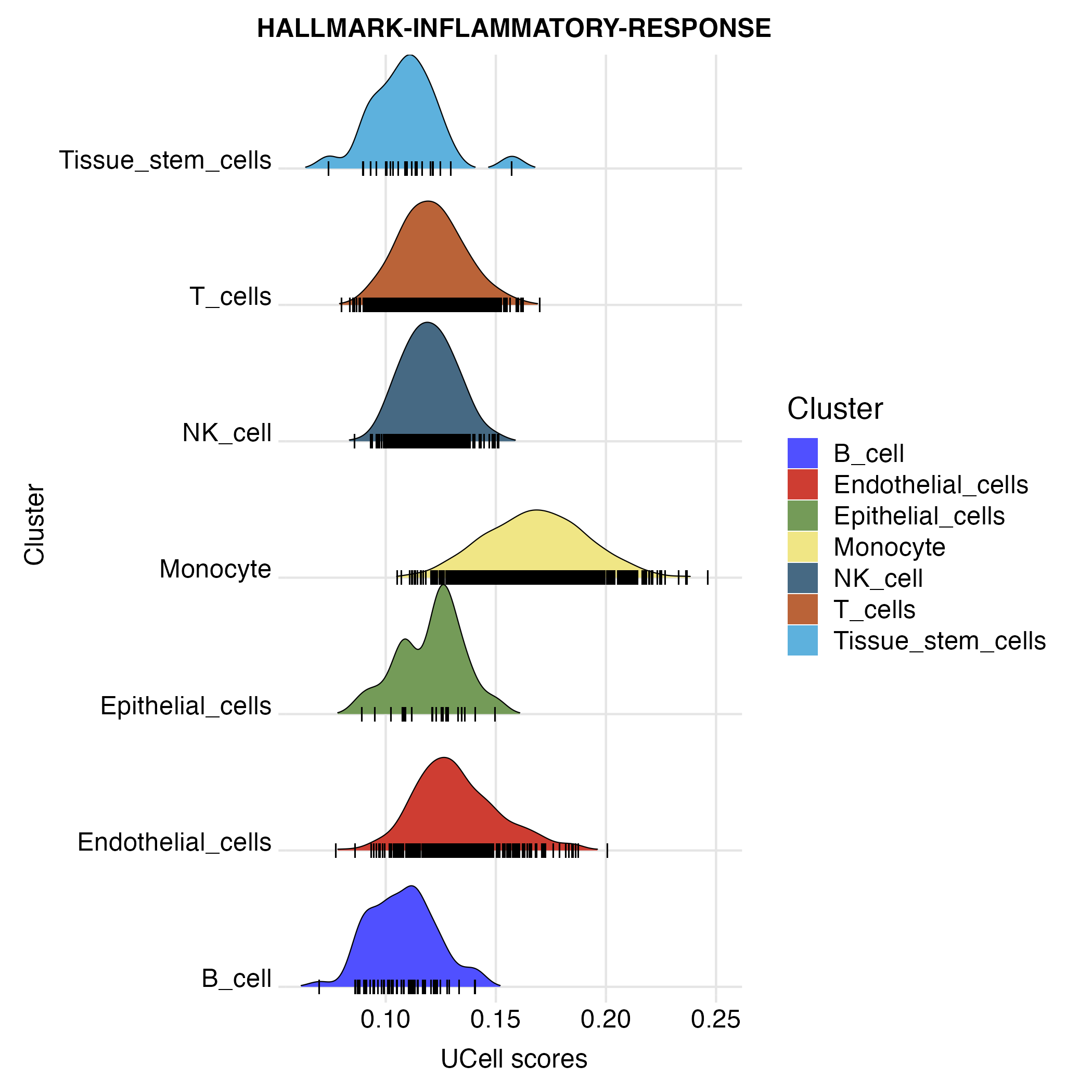
densityheatmap <- irGSEA.densityheatmap(object = scRNA,method = "UCell",show.geneset = "HALLMARK-INFLAMMATORY-RESPONSE")