@agpwhy
2022-02-20T13:13:02.000000Z
字数 4289
阅读 378
王胖的生信笔记第38期:螺旋图
大家不知道有没有见过这种图
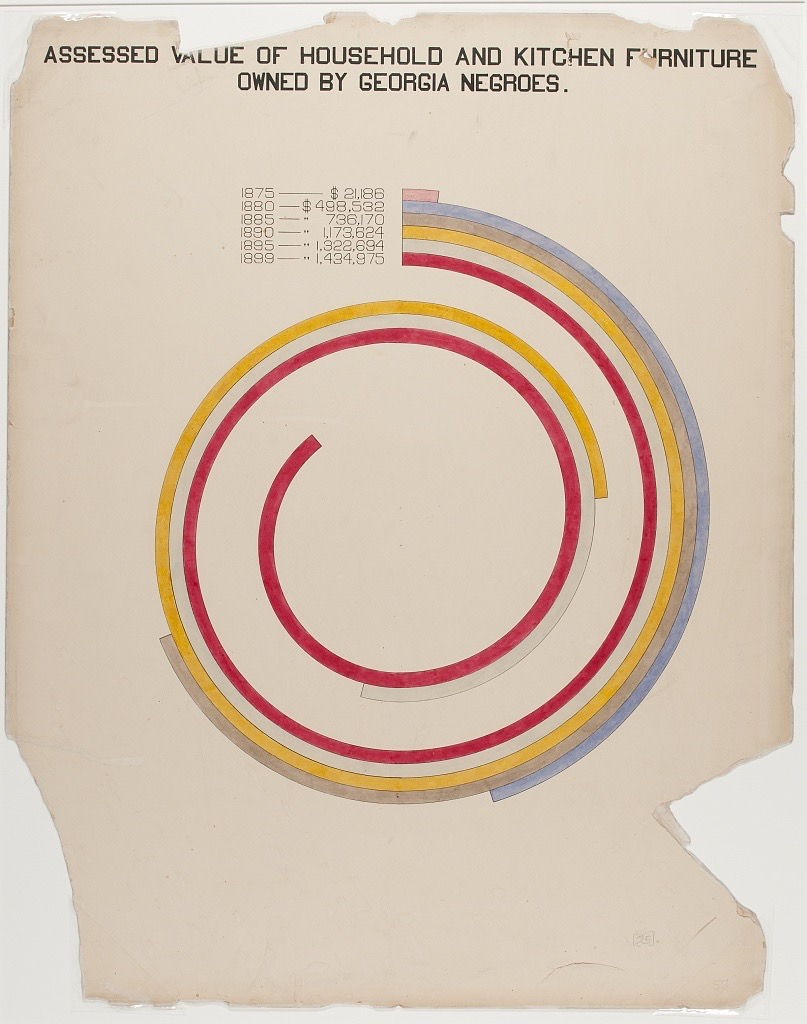
就是数据一圈一圈盘绕一起来的。这可以算做柱状图的一种变种。好处在于可以节约空间,保证可以调整尺寸。这张图使用decksh做的,这个我不会,也觉得为了做个图专门去学习一个完全新的工具有点不划算。
于是搜索了一下,Gu ZuGuang老师开发了个R包,就是用来做这样的图的。
https://github.com/jokergoo/spiralize
也有写教程,这里就把教程里摘取一些简单写写。
怎么做螺旋图
最主要的是要理解坐标系的一个变换。
我们最常见到的是x轴,y轴这样的坐标系,也就是笛卡尔坐标系。但是螺旋的图需要的是极坐标系,这个应该是高中数学知识,不过可能很多人都有点忘记了。没关系,不用会数学,大概要知道一下这个概念就行。
spiral_initialize(start = 90, end = 360)spiral_track()
这个就是从90度的地方开始,360度的地方结束。

多绕几圈也可以,就修改一下结束的度数(可以大于360)。

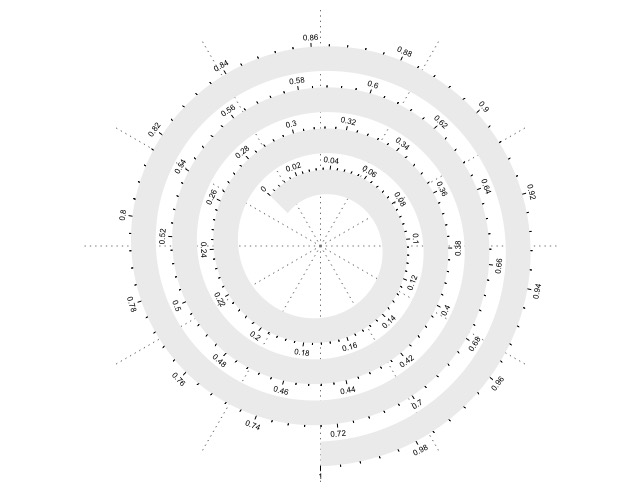
这个顺时针,逆时针都可以选择(默认的按照极坐标系是逆时针的)。也可以加上坐标轴。
spiral_initialize(start = 135 + 360, clockwise = TRUE)spiral_track(height = 0.6)spiral_axis()
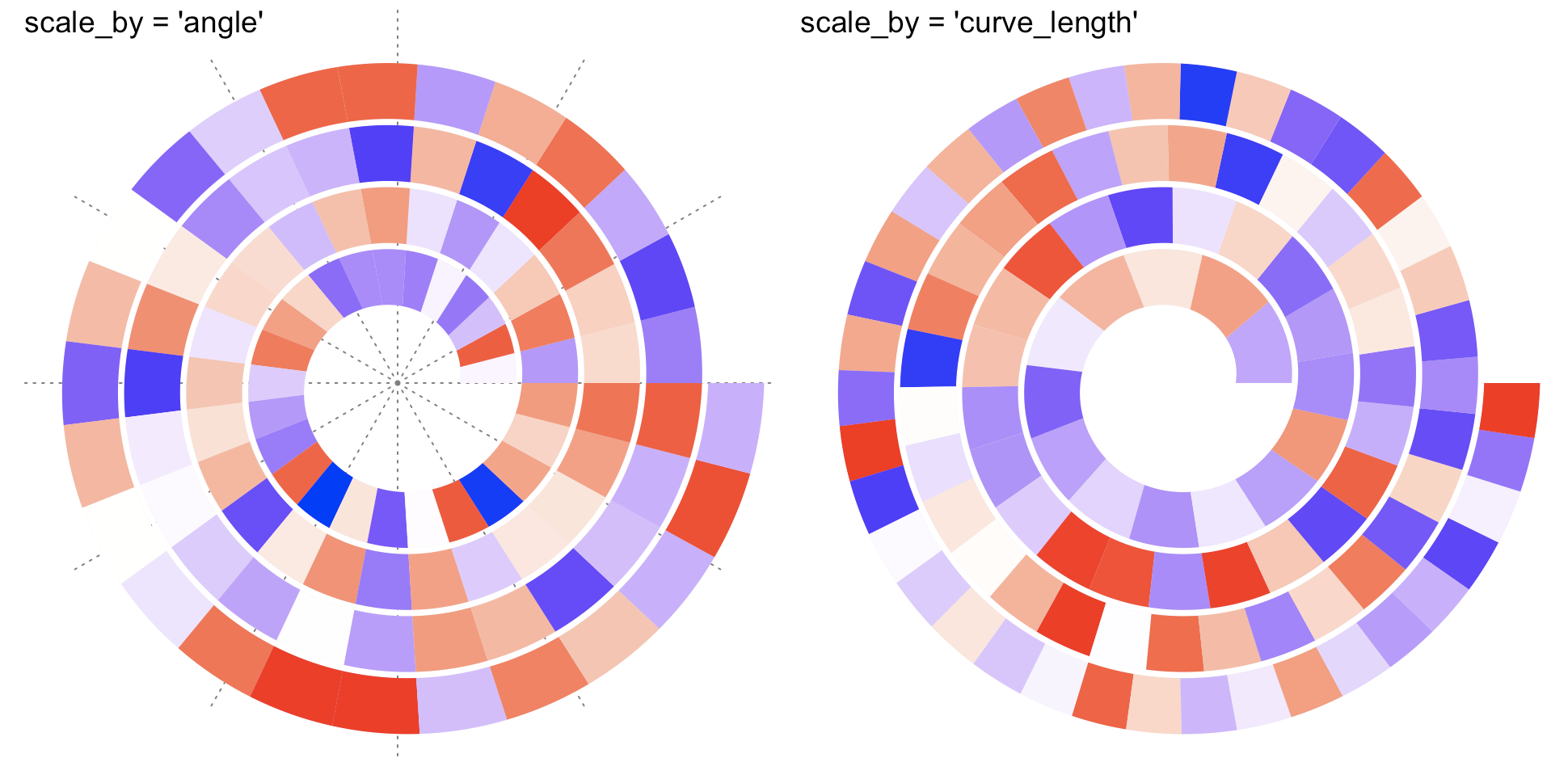
还有一个可以选择的选项就是做报表的单位是以角度为准还是以弧长为准(这也是中学数学概念,不严谨来说就是标注的单位是以角度还是长度为准)。
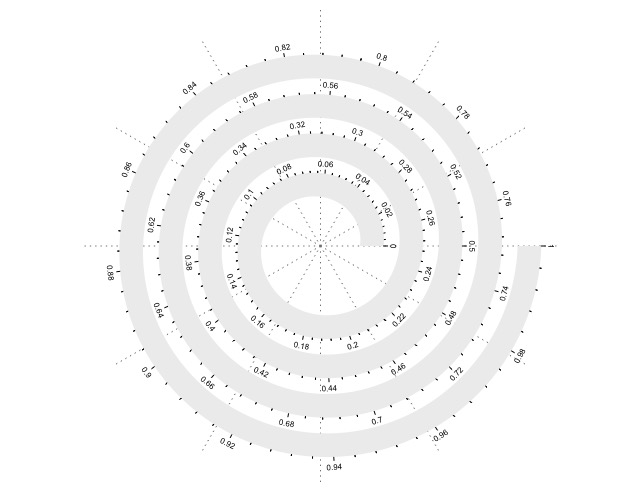
spiral_initialize(scale_by = "angle") # 默认是角度为准spiral_track(height = 0.6)spiral_axis()

spiral_initialize(scale_by = "curve_length")(弧长为主)spiral_track(height = 0.6)spiral_axis()
这是以弧长为主。
节律数据的例子
教程里讲了好多。我拿两个基础医学实验上可能比较常用的例子简单摘录一下。
这个是Elf1基因在WT/KO(Bmal1)两组小鼠中表达随时间变化的区别。每一圈是一天(3小时测一次,一共测了3天)。这是不是就很适合用过螺旋图?
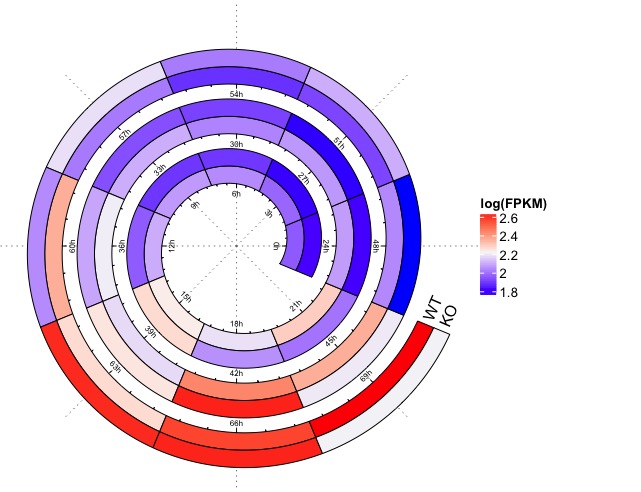
# 读取示范数据expr = readRDS(system.file("extdata", "circadian_expression_mouse_Elf1.rds", package = "spiralize"))# 设置时间节点t = seq(0, 69, by = 3)rg = range(expr)# 开始准备做热图配色col = colorRamp2(c(rg[1], mean(rg), rg[2]), c("blue", "#F7F7F7", "red"))# 铺设螺旋轴spiral_initialize(xlim = c(0, 69), period = 24, polar_lines_by = 45,vp_param = list(x = unit(0, "npc"), just = "left"))# 设置刻度spiral_track(ylim = c(0, 1), height = 0.7)spiral_rect(t - 1.5, 0, t + 1.5, 0.5, gp = gpar(fill = col(expr[1:24])))spiral_rect(t - 1.5, 0.5, t + 1.5, 1, gp = gpar(fill = col(expr[1:24 + 24])))spiral_axis(at = t, labels = paste0(t, "h"), h = "bottom")# 加上分组文字标注spiral_text(69 + 1.6, 0.25, "WT", just = "left", facing = "outside")spiral_text(69 + 1.6, 0.75, "KO", just = "left", facing = "outside")# 加上图注lgd = Legend(title = "log(FPKM)", col_fun = col)draw(lgd, x = unit(1, "npc") + unit(2, "mm"), just = "left")
表观遗传的例子
用的是UCSD数据上BCAT2这个基因在肺组织里甲基化以及组蛋白乙酰化的展示。如果是一长条展示,排版就很烦,但是呢,用螺旋图就能用一个方块区域,方便排版了。
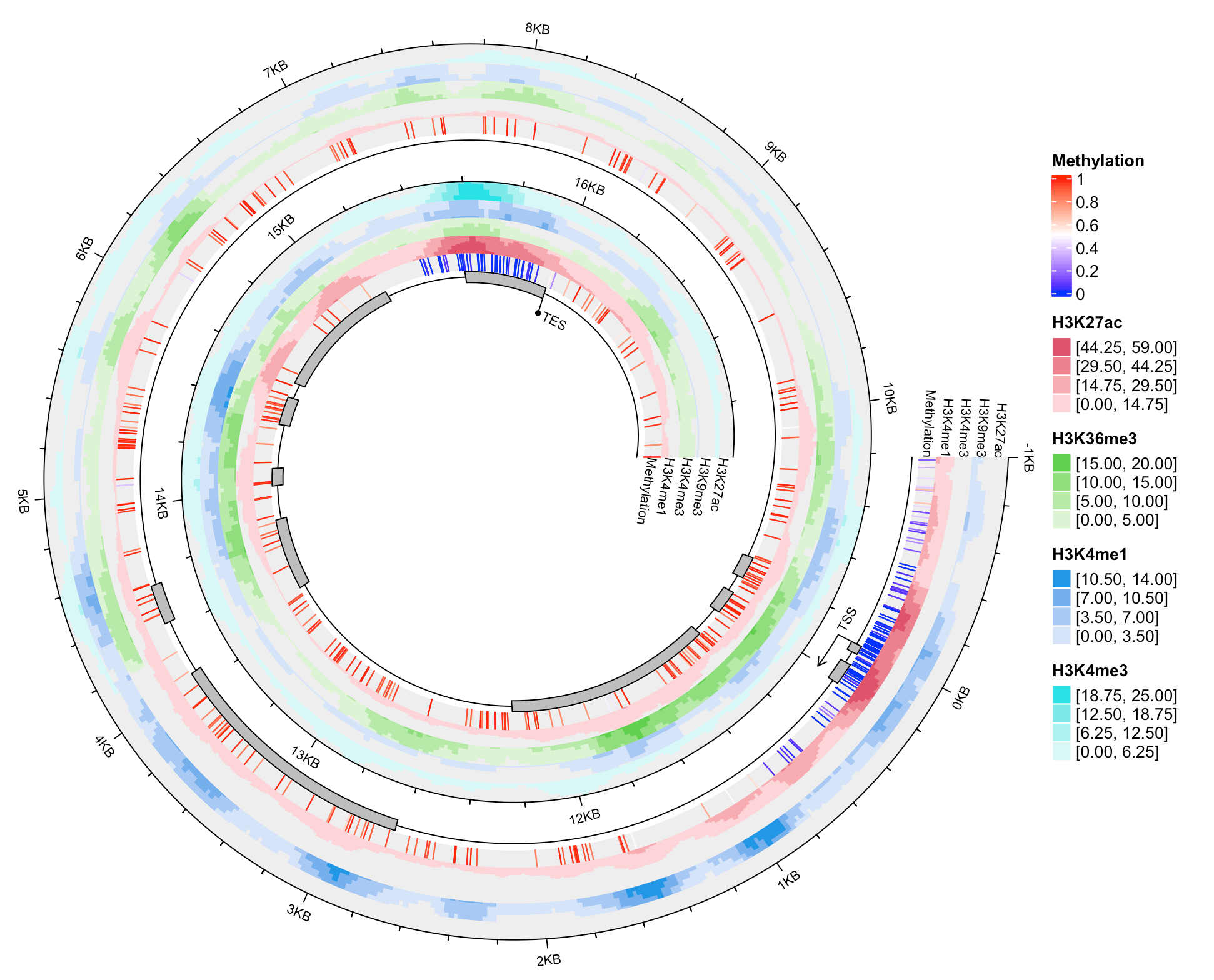
这个有点复杂。我讲不太来。
df_list = readRDS(system.file("extdata", "BCAT2_tracks.rds", package = "spiralize"))spiral_initialize_by_gcoor(xlim = c(df_list$range[1, 2], df_list$range[1, 3]), end = 360*3,vp_param = list(x = unit(0, "npc"), just = "left"))exons = df_list$exonsexons = exons[exons[, 8] == "exon", ]spiral_track(height = 0.1, background = FALSE)spiral_lines(TRACK_META$xlim, 0.5)spiral_rect(exons[, 2], 0.1, exons[, 3], 0.9, gp = gpar(fill = "grey"))spiral_segments(49314286, 0, 49314286, -1)spiral_segments(49314286, -1, 49314286 - 200, -1, arrow = arrow(length = unit(2, "mm")))spiral_text(49314286, -1, "TSS", hjust = -0.2, facing = "outside", gp = gpar(fontsize = 8))spiral_segments(49298318, 0, 49298318, -1)spiral_text(49298318, -1, "TES", hjust = -0.2, facing = "inside", gp = gpar(fontsize = 8))spiral_points(49298318, -1, pch = 16)meth = df_list$methspiral_track(height = 0.13)meth_col = colorRamp2(c(0, 0.5, 1), c("blue", "white", "red"))spiral_segments(meth[, 2], 0, meth[, 2], 1, gp = gpar(col = meth_col(meth[, 5])))lgd_list = list(Legend(title = "Methylation", col_fun = meth_col))i = 1for(mark in grep("H3", names(df_list), value = TRUE)) {df = df_list[[mark]]i = i + 1spiral_track(height = 0.13)lt = spiral_horizon( (df[, 2] + df[, 3])/2, df[, 5], pos_fill = i,use_bars = TRUE, bar_width = df[, 3] - df[, 2])lgd_list = c(lgd_list, list(horizon_legend(lt, title = mark)))}spiral_lines(TRACK_META$xlim, 1)at = seq(df_list$range[1, 3], df_list$range[1, 2], by = -1000)labels = -(seq(df_list$range[1, 3], df_list$range[1, 2], by = -1000) - 49314286)labels = paste0(labels/1000, "KB")labels[labels == "11KB"] = ""spiral_axis(at = at, labels = labels, labels_gp = gpar(fontsize = 8))spiral_text(49297318, 0.5, "Methylation", hjust = 0, facing = "inside", gp = gpar(fontsize = 8), track_index = 2)spiral_text(49297318, 0.5, "H3K4me1", hjust = 0, facing = "inside", gp = gpar(fontsize = 8), track_index = 3)spiral_text(49297318, 0.5, "H3K4me3", hjust = 0, facing = "inside", gp = gpar(fontsize = 8), track_index = 4)spiral_text(49297318, 0.5, "H3K9me3", hjust = 0, facing = "inside", gp = gpar(fontsize = 8), track_index = 5)spiral_text(49297318, 0.5, "H3K27ac", hjust = 0, facing = "inside", gp = gpar(fontsize = 8), track_index = 6)spiral_text(49315286, 0.5, "Methylation", hjust = 1, facing = "inside", gp = gpar(fontsize = 8), track_index = 2)spiral_text(49315286, 0.5, "H3K4me1", hjust = 1, facing = "inside", gp = gpar(fontsize = 8), track_index = 3)spiral_text(49315286, 0.5, "H3K4me3", hjust = 1, facing = "inside", gp = gpar(fontsize = 8), track_index = 4)spiral_text(49315286, 0.5, "H3K9me3", hjust = 1, facing = "inside", gp = gpar(fontsize = 8), track_index = 5)spiral_text(49315286, 0.5, "H3K27ac", hjust = 1, facing = "inside", gp = gpar(fontsize = 8), track_index = 6)lgd = packLegend(list = lgd_list, max_height = unit(8, "in"))draw(lgd, x = unit(1, "npc") + unit(2, "mm"), just = "left")
