@agpwhy
2022-01-05T03:25:28.000000Z
字数 2402
阅读 1026
王胖的生信笔记第32期:基于芯片实战的双向富集
做完芯片数据下载和简单控制后,后续比较常见生信分析之一就是做富集分析。富集可以基于各种公共数据,包括GO,KEGG,Reactome等等。
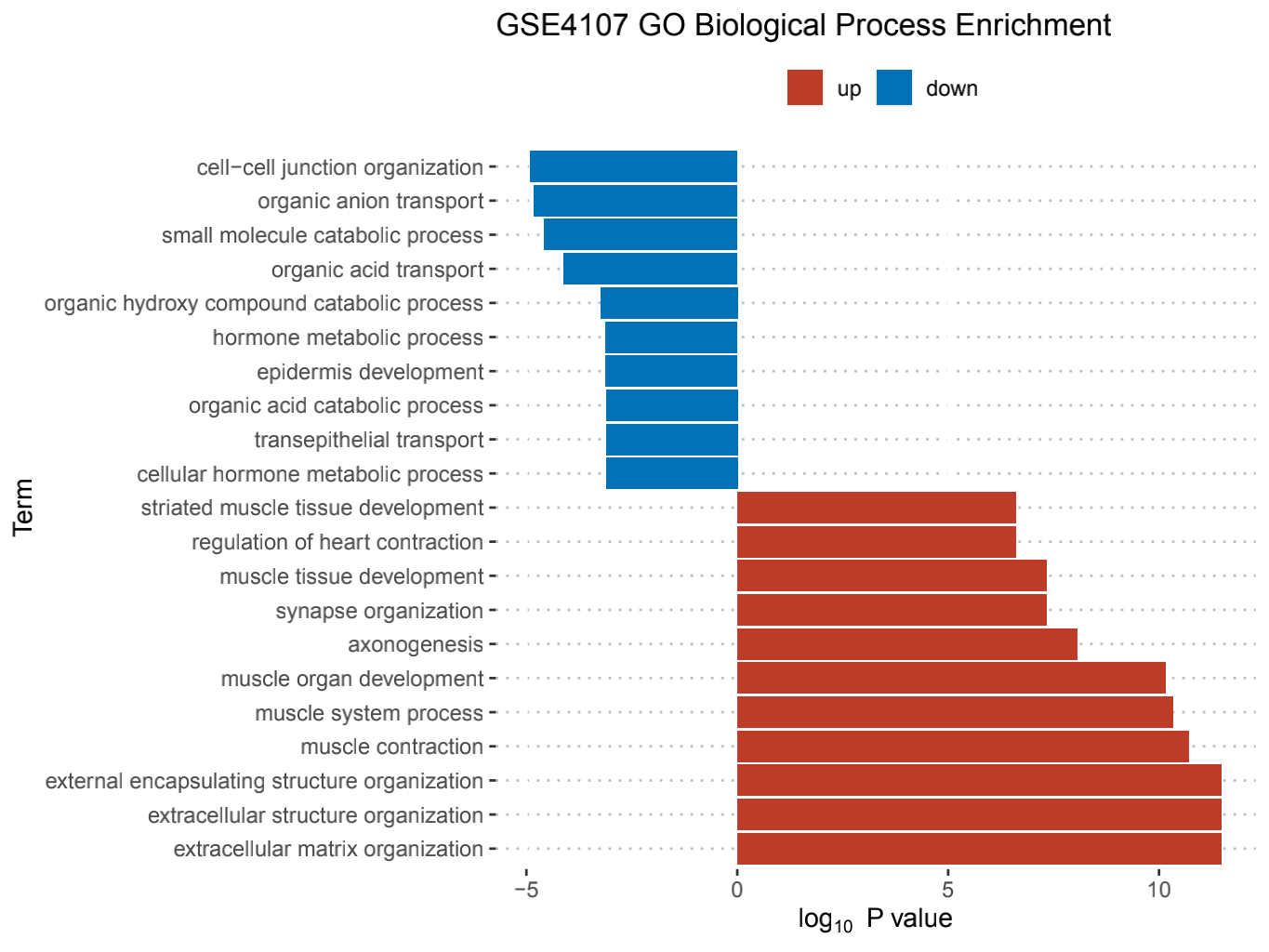
大家可能对传统的富集已经有办法了,怎么样作出像这样双向富集的图呢?(这个跟着生信技能树即可)
当然这个做的比较简单。后面我们来一步步做。
准备环境
library(stringr)library(AnnoProbe)library(tinyarray)library(tidyverse)rm(list = ls())options(stringsAsFactors = F)
下载数据
gse = "GSE4107"geo = geo_download(gse)geo$exp = log(geo$exp+1)change_list = ifelse(str_detect(geo$pd$title,"control"),"control","treat")change_list = factor(change_list,levels = c("control","treat"))
ID转化+一步傻瓜化数据处理
find_anno(geo$gpl)ids <- AnnoProbe::idmap('GPL570')dcp = get_deg_all(geo$exp,change_list,ids,logFC_cutoff = 1,scale_before = T,cluster_cols = F)
到了这一步就已经完成了数据准备了。后面就是利用这个数据做筛选变化,作出我们需要的图。
数据筛选,分开上调下调显著基因
因为我们要做上调下调两个方向的富集,所以要分开两个基因集。比较好的是用Tinyarray这个包还是有懒人法一步可以到位。
deg <- filter(dcp[[1]],adj.P.Val<0.05)# 如果要看具体数值,可以这里筛选来看degup <- dcp[[2]]$up$upgenesdegdown <- dcp[[2]]$down$downgeness
如果你用的是和我一样的数据和步骤,可以看到这样的结果。
head(degup)
[1] "FOSB" "FOS" "DUSP1" "CCDC3" "EGR1" "RERGL"
head(degdown)
[1] "NR1H4" "SLC51A" "BCKDHB" "MEP1B" "ETNK1" "AKR1C3"
分开富集
ego_ALL_up <- enrichGO(gene = degup,OrgDb = 'org.Hs.eg.db',keyType = 'SYMBOL',ont = "BP",pAdjustMethod = "BH",pvalueCutoff = 0.01,qvalueCutoff = 0.05)ego_all_up <- data.frame(ego_ALL_up)ego_all_up$direction <- "up"ego_ALL_down <- enrichGO(gene = degdown,OrgDb = 'org.Hs.eg.db',keyType = 'SYMBOL',ont = "BP",pAdjustMethod = "BH",pvalueCutoff = 0.01,qvalueCutoff = 0.05)ego_all_down <- data.frame(ego_ALL_down)ego_all_down$direction <- "down"
分别选取上下调最显著通路再合并
ego_all <- rbind(ego_all_up,ego_all_down)ego_all_up_sub <- ego_all_up %>% top_n(n = 10, wt = -`p.adjust`)ego_all_down_sub <- ego_all_down %>% top_n(n = 10, wt = -`p.adjust`)ego_all_sub <- rbind(ego_all_up_sub,ego_all_down_sub)ego_all_sub$Description <- substring(ego_all_sub$Description,1,70)ego_all_sub$pvalue = -log10(ego_all_sub$p.adjust)
开始作图
ego_all_sub[ego_all_sub$direction %in% "down",]$pvalue <- ego_all_sub[ego_all_sub$direction %in% "down",]$pvalue*-1ego_all_sub[duplicated(ego_all_sub$Description),"Description"] <- paste0(ego_all_sub[duplicated(ego_all_sub$Description),"Description"],".1")ego_all_sub$Description <- factor(ego_all_sub$Description,levels = ego_all_sub[order(ego_all_sub$pvalue,decreasing = T),"Description"])ego_all_sub$direction <- factor(ego_all_sub$direction,levels=c("up","down"))ggplot(ego_all_sub,aes(x=Description,y=pvalue, fill=factor(direction)))+geom_bar(stat="identity")+coord_flip( )+scale_fill_nejm()+theme_pubclean()+labs(x = "", y = "-log10 P Value",title = "GO Biological Process Enrichment")
完成就是这样啦。
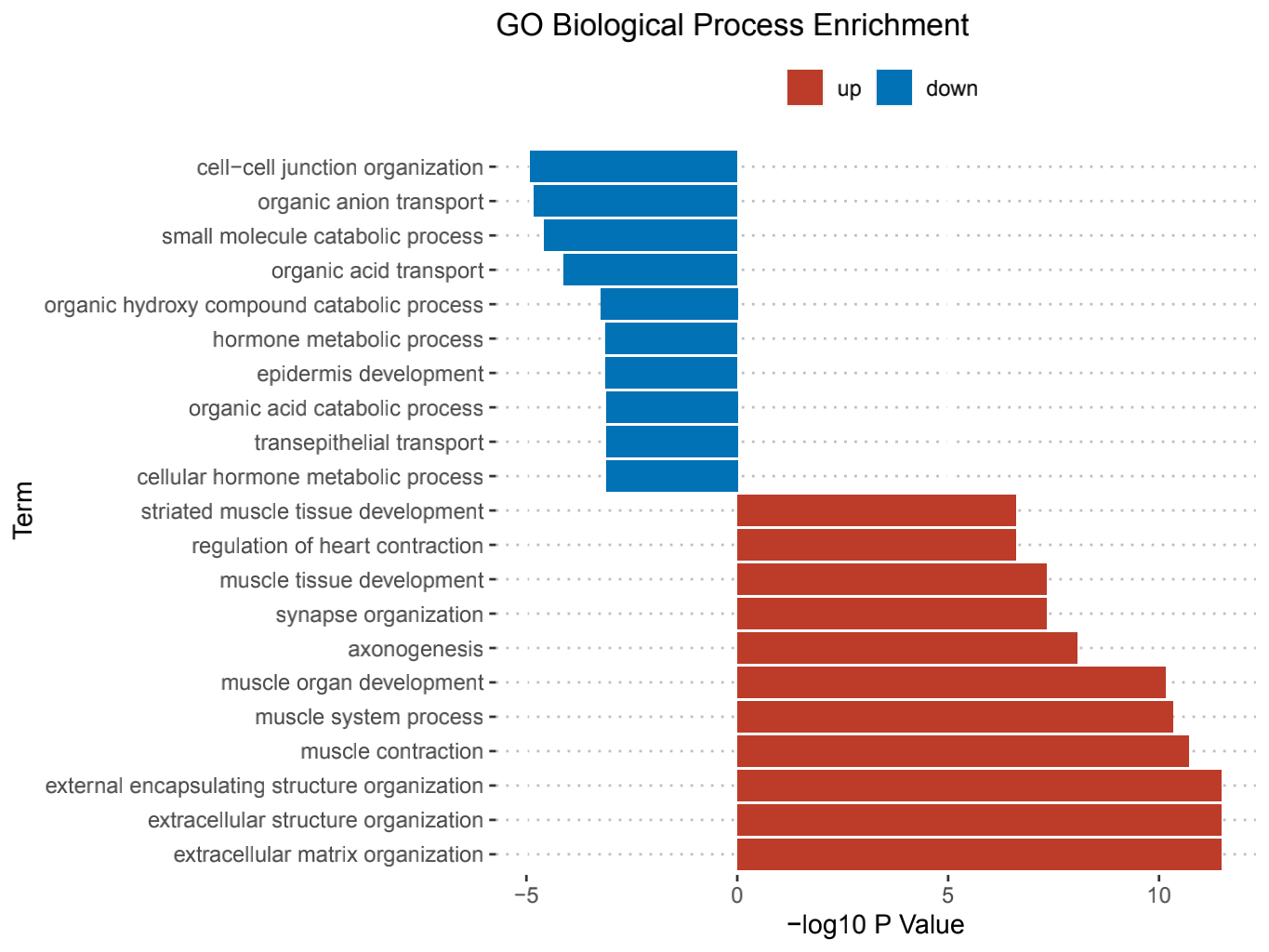
其实还有很多地方可以调整。